Summary: The Gini Index is calculated by subtracting the sum of the squared probabilities of each class from one. It favors larger partitions. Information Gain multiplies the probability of the class times the log (base=2) of that class probability. Information Gain favors smaller partitions with many distinct values. Ultimately, you have to experiment with your data and the splitting criterion.
[table]
Algo / Split Criterion, Description, Tree Type
Gini Split / Gini Index, Favors larger partitions. Very simple to implement., CART
Information Gain / Entropy, Favors partitions that have small counts but many distinct values., ID3 / C4.5
[/table]
We’ll talk about two splitting criteria in the context of R’s rpart library. It’s important to experiment with different splitting methods and to analyze your data before you commit to one method. Each splitting algorithm has its own bias which we’ll briefly explore. The code is available on GitHub here.
Using Gini Split / Gini Index
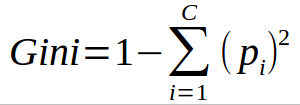
- Favors larger partitions.
- Uses squared proportion of classes.
- Perfectly classified, Gini Index would be zero.
- Evenly distributed would be 1 – (1/# Classes).
- You want a variable split that has a low Gini Index.
- The algorithm works as 1 – ( P(class1)^2 + P(class2)^2 + … + P(classN)^2)
The Gini index is used in the classic CART algorithm and is very easy to calculate.
Gini Index:
for each branch in split:
Calculate percent branch represents #Used for weighting
for each class in branch:
Calculate probability of class in the given branch.
Square the class probability.
Sum the squared class probabilities.
Subtract the sum from 1. #This is the Ginin Index for branch
Weight each branch based on the baseline probability.
Sum the weighted gini index for each split.
Here’s that same process in R as a function. It takes advantage of R’s ability to calculate sums and probabilities quickly with the prop.table function. The only difference between the pseudo-code above and the actual code is that I take into account the Null situation where we don’t specify a split variable.
gini_process <-function(classes,splitvar = NULL){
#Assumes Splitvar is a logical vector
if (is.null(splitvar)){
base_prob <-table(classes)/length(classes)
return(1-sum(base_prob**2))
}
base_prob <-table(splitvar)/length(splitvar)
crosstab <- table(classes,splitvar)
crossprob <- prop.table(crosstab,2)
No_Node_Gini <- 1-sum(crossprob[,1]**2)
Yes_Node_Gini <- 1-sum(crossprob[,2]**2)
return(sum(base_prob * c(No_Node_Gini,Yes_Node_Gini)))
}
With all of this code, we can apply this on a set of data.
data(iris) gini_process(iris$Species) #0.6667 gini_process(iris$Species,iris$Petal.Length<2.45) #0.3333 gini_process(iris$Species,iris$Petal.Length<5) #0.4086 gini_process(iris$Species,iris$Sepal.Length<6.4) #0.5578
In this case, we would choose the Petal.Length<2.45 as the optimal variable / condition to split on. It has the lowest Gini Index.
Splitting with Information Gain and Entropy
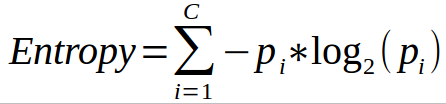
- Favors splits with small counts but many unique values.
- Weights probability of class by log(base=2) of the class probability
- A smaller value of Entropy is better. That makes the difference between the parent node’s entropy larger.
- Information Gain is the Entropy of the parent node minus the entropy of the child nodes.
- Entropy is calculated [ P(class1)*log(P(class1),2) + P(class2)*log(P(class2),2) + … + P(classN)*log(P(classN),2)]
When you use Information Gain, which uses Entropy as the base calculation, you have a wider range of results. The Gini Index caps at one. The maximum value for Entropy depends on the number of classes. It’s based on base-2, so if you have…
- Two classes: Max entropy is 1.
- Four Classes: Max entropy is 2.
- Eight Classes: Max entropy is 3.
- 16 classes: Max entropy is 4.
With that being said, let’s take a look at how you might calculate Entropy.
Entropy:
for each branch in split:
Calculate percent branch represents #Used for weighting
for each class in branch:
Calculate probability of class in the given branch.
Multiply probability times log(Probability,base=2)
Multiply that product by -1
Sum the calculated probabilities.
Weight each branch based on the baseline probability.
Sum the weighted entropy for each split.
It’s very similar to the Gini Index calculation. The only real difference is what you do with the class probabilities. Here it is again in R.
info_process <-function(classes,splitvar = NULL){
#Assumes Splitvar is a logical vector
if (is.null(splitvar)){
base_prob <-table(classes)/length(classes)
return(-sum(base_prob*log(base_prob,2)))
}
base_prob <-table(splitvar)/length(splitvar)
crosstab <- table(classes,splitvar)
crossprob <- prop.table(crosstab,2)
No_Col <- crossprob[crossprob[,1]>0,1]
Yes_Col <- crossprob[crossprob[,2]>0,2]
No_Node_Info <- -sum(No_Col*log(No_Col,2))
Yes_Node_Info <- -sum(Yes_Col*log(Yes_Col,2))
return(sum(base_prob * c(No_Node_Info,Yes_Node_Info)))
}
Again, we can run this set of code against our data.
data(iris) info_process(iris$Species) #1.584963 info_process(iris$Species,iris$Petal.Length<2.45) #0.6666667 info_process(iris$Species,iris$Petal.Length<5) #0.952892 info_process(iris$Species,iris$Sepal.Length<6.4) #1.302248
The difference between the parent Entropy and the Petal.Length of less than 2.45 is the greatest (1.58-0.667) so it’s still the most important variable.
Information Gain and Gini Index by Hand
Let’s walk through an example of calculating a few nodes.
[table]Class,Var1,Var2,
A,0,33,
A,0,54,
A,0,56,
A,0,42,
A,1,50,
B,1,55,
B,1,31,
B,0,-4,
B,1,77,
B,0,49[/table]
- We’re trying to predict the class variable.
- For numeric variables, you would go from distinct value to distinct value and check the split as less than and greater than or equal to.
We’ll first try using Gini Index on a couple values. Let’s try Var1 == 1 and Var2 >=32.
Gini Index Example: Var1 == 1
- Baseline of Split: Var1 has 4 instances (4/10) where it’s equal to 1 and 6 instances (6/10) when it’s equal to 0.
- For Var1 == 1 & Class == A: 1 / 4 instances have class equal to A.
- For Var1 == 1 & Class == B: 3 / 4 instances have class equal to B.
- Gini Index here is 1-((1/4)^2 + (3/4)^2) = 0.375
- For Var1 == 0 & Class== A: 4 / 6 instances have class equal to A.
- For Var1 == 0 & Class == B: 2 / 6 instances have class equal to B.
- Gini Index here is 1-((4/6)^2 + (2/6)^2) = 0.4444
- We then weight and sum each of the splits based on the baseline / proportion of the data each split takes up.
- 4/10 * 0.375 + 6/10 * 0.444 = 0.41667
Gini Index Example: Var2 >= 32
- Baseline of Split: Var2 has 8 instances (8/10) where it’s equal >=32 and 2 instances (2/10) when it’s less than 32.
- For Var2 >= 32 & Class == A: 5 / 8 instances have class equal to A.
- For Var2 >= 32 & Class == B: 3 / 8 instances have class equal to B.
- Gini Index here is 1-((5/8)^2 + (3/8)^2) = 0.46875
- For Var2 < 32 & Class == A: 0 / 2 instances have class equal to A.
- For Var2 < 32 & Class == B: 2 / 2 instances have class equal to B.
- Gini Index here is 1-((0/2)^2 + (2/2)^2) = 0
- We then weight and sum each of the splits based on the baseline / proportion of the data each split takes up.
- 8/10 * 0.46875 + 2/10 * 0 = 0.375
Based on these results, you would choose Var2>=32 as the split since its weighted Gini Index is smallest. The next step would be to take the results from the split and further partition. Let’s take the 8 / 10 records and try working with an Information Gain Split.
[table]
Class,Var1,Var2,
A,0,33,
A,0,54,
A,0,56,
A,0,42,
A,1,50,
B,1,55,
B,1,77,
B,0,49[/table]
Information Gain Example: Var2<45.5
Again, we’ll follow a similar procedure.
- Baseline of Split: Var2 has 2 instances (2/8) where it’s < 45.5 and 6 instances (6/8) when it’s >=45.5.
- For Var2 < 45.5 & Class == A: 2 / 2 instances have class equal to A.
- For Var2 < 45.5 & Class == B: 0 / 2 instances have class equal to B.
- Entropy here is -1 * ((2/2)*log(2/2, 2)) = 0
- Notice how class B isn’t represented here at all.
- For Var2 >= 45.5 & Class == A: 3 / 6 instances have class equal to A.
- For Var2 >= 45.5 & Class == B: 3 / 6 instances have class equal to B.
- Entropy here is -1 * ((3/6)*log(3/6, 2) +(3/6)*log(3/6, 2)) = 1
- We then weight and sum each of the splits based on the baseline / proportion of the data each split takes up.
- 2/8 * 0 + 6/8 * 1 = 0.75
Information Gain Example: Var2<65.5
- Baseline of Split: Var2 has 7 instances (7/8) where it’s < 65.5 and 1 instance (1/8) when it’s >=65.5.
- For Var2 < 65.5 & Class == A: 5 / 7 instances have class equal to A.
- For Var2 < 65.5 & Class == B: 2 / 7 instances have class equal to B.
- Entropy here is -1 * ((5/7)*log(5/7, 2) +(2/7)*log(2/7, 2)) = 0.8631
- For Var2 >= 65.5 & Class == A: 0 / 1 instances have class equal to A.
- For Var2 >= 65.5 & Class == B: 1 / 1 instances have class equal to B.
- Entropy here is -1 * ((1/1)*log(1/1, 2)) = 0
- Notice how class A isn’t represented here at all.
- We then weight and sum each of the splits based on the baseline / proportion of the data each split takes up.
- 7/8 * 0.8631 + 1/8 * 0 = 0.7552
Based on Information Gain, we would choose the split that has the lower amount of entropy (since it would maximize the gain in information). We would choose Var2 < 45.5 as the next split to use in the decision tree.
As an exercise for you, try computing the Gini Index for these two variables. You should see that we would choose Var2 < 65.5!
When Information Gain and Gini Index Choose Different Variables
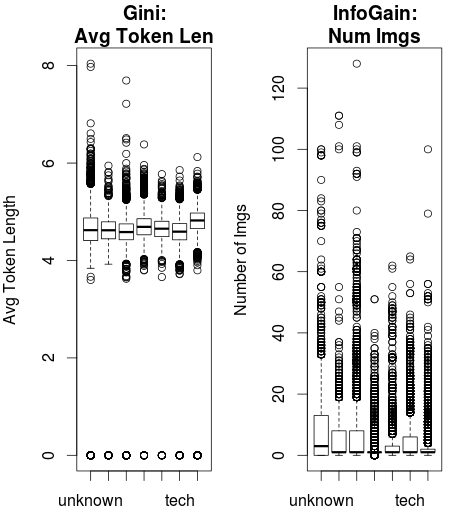
The differences are much more intuitive when you look at some real data and how the splitting method would make an impact.
This data from the UCI Machine Learning Repository shows the popularity of webpages from Mashable. Two variables, Average Token Length and Number of Images are entered into a classification decision tree.
Using Gini Index as the splitting criteria, Average Token Length is the root node.
Using Information Gain, Number of Images is selected as the root node.
You can see the relatively tighter spread of the Average Token Length and the wider dispersion of the Number of Images.
Ultimately, the choice you make comes down to examining your data and being aware of the biases of your algorithms. Again, the code for this example is available on GitHub here.